Learning from evolution: Using AI language models to design functional artificial proteins
Proteins are the essential workhorses of biology that carry out spectacularly varied functions from fighting viruses in the human body to degrading plastics. Fundamentally, proteins are chains of amino acid molecules that have evolved naturally over billions of years through mutation. Instead of relying on the long arc of natural evolution, what if we could take control and design proteins ourselves from scratch?
In this study, we look to artificial intelligence (AI) for help and build on the success of AI language models in generating highly realistic natural language sentences. We show that our language model, named ProGen, can learn the language of proteins to generate artificial protein sequences across multiple families. We validate our model by synthesizing artificial antibacterial lysozyme proteins in the lab and compare their antibacterial strength to natural proteins. Even though many of our artificial proteins are quite different from natural proteins, they are just as effective in antibacterial performance. Our work demonstrates we can use AI as a controllable tool to design proteins for our intended purposes in biology. We mark this milestone by reporting the first known 3D structure of an artificial protein designed fully by AI. We believe a new field of synthetic biology with generative AI has emerged and can lead to solutions for challenges in human disease and the environment. Please refer to our paper for full details or contact us at progen@salesforce.com
Summary: We show that AI can learn the language of biology to create artificial proteins across multiple protein types that are functional and unseen in nature.
Protein Evolution and Design
Designing functional artificial proteins is a grand challenge in biology. In fact, the design space of possible proteins is greater than the number of atoms in the entire universe! An important scientific method for designing proteins is “Directed Evolution”, a Nobel prize-winning technique that works by imitating natural evolution in a laboratory. However, evolving proteins from scratch in a laboratory can be prohibitively slow or in many cases, not work at all. Our goal is to circumvent the need for evolution and design proteins with desired properties from scratch.
So how do we learn to design proteins? We take advantage of a key trend in biology: the exponential growth in publicly-available raw protein sequence data, enabled by the dramatic reduction in sequencing costs. Datasets containing millions of protein sequences can be used by AI models to learn nature’s blueprint for proteins. We’ve recently witnessed impressive prediction performance by training AI models with access to millions of sequences, like with structure prediction tasks by AlphaFold. We, however, aim to go beyond predicting the structure of a given protein. Our goal is to create artificial proteins from scratch that are useful and functional. We make strides toward a general-purpose protein design system that can generate protein sequences for different protein families through the usage of a class of AI models called neural language models, described next.
ProGen: Learning the Language of Proteins with AI
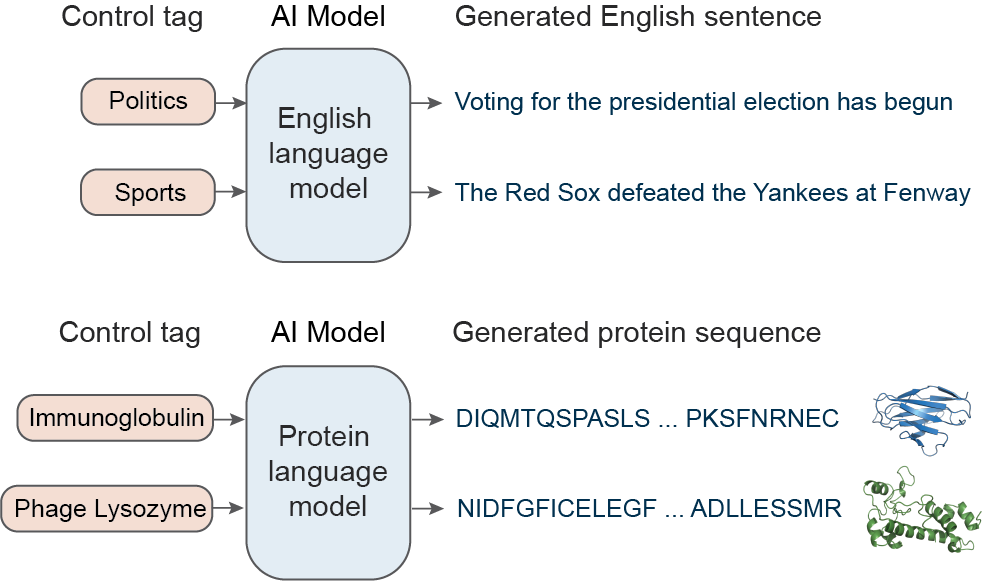
AI algorithms called “neural language models” have shown remarkable success in generating artificial text by learning to imitate human language. If trained on enough data, language models are able to generate novel text that is indistinguishable from human-written text! A key insight for our work is that proteins can be represented as a language made up of amino acids, the 20 molecules that make up every protein. In the same way that words are strung together one-by-one to form text sentences, amino acids are strung together one-by-one to make proteins. Building on this insight, we apply neural language modeling to proteins for generating realistic, yet novel protein sequences.
Specifically, we train a “conditional” language model, which is a type of model that can be steered with user inputs to generate language with certain user-specified properties called “control tags.” In the case of human language, these control tags may be properties like style, topics, or dates. For instance, if you give a conditional language model a control tag that says “Politics”, it will likely generate a sentence about a political topic, like elections. For proteins, the control tags are biological properties such as protein family, biological process, or molecular function. So if you give a conditional language model a control tag that specifies a protein family (e.g. “Phage Lysozyme”, an antibacterial protein), it will likely generate a protein with a sequence of amino acids within the Phage Lysozyme family. Conditional language models allow for significantly more control over what types of sequences are generated, making them more useful for designing proteins with specific properties.
In prior work, we trained a conditional language model, called ProGen, on 280 million protein sequences from protein databases. Our previous blog post focused on the machine learning techniques in natural language processing (NLP) that are useful for modeling proteins. Our simulated experiments showed that ProGen had learned rules on how to generate sequences of amino acids in a structurally-viable manner. This is analogous to AI language models trained on human languages capturing word usage patterns and grammar rules. However, the gold standard for evaluating ProGen would be to show that a sequence of amino acids generated by ProGen can be synthesized in the real-world and perform its intended function. In the current study, we take this significant next step.
Laboratory Evaluation of ProGen-generated Antibacterial Proteins
To test artificial proteins generated by ProGen, we chose antibacterial proteins called lysozymes. Lysozymes, which were the first antibiotic ever discovered, are incredibly diverse, have several evolutionary families, and are even present in our tears and mucus. Each family of lysozyme is structurally different, meaning that the proteins fold into different shapes. We selected five specific families of lysozymes for generation, which contain proteins that are 90-180 amino acid-long on average. To improve generation quality, we trained ProGen further on a publicly-available database of natural lysozymes. We then used control tags to tell the model to generate artificial proteins from these five lysozyme families.
Our experiments pit artificial proteins against natural proteins in the lab. We worked with Tierra Biosciences and Professor James Fraser’s lab at UCSF to make this a reality. First, scientists at Tierra Biosciences synthesized natural and artificial proteins by repurposing the cell’s complex machinery as a factory for creating proteins from custom-printed DNA. The proteins were tested for antibacterial function in what’s known as an activity assay. Lysozymes can kill certain types of bacteria by triggering a reaction that dissolves bacterial cell walls. The activity assay enables characterization of this antibacterial reaction by emitting light (i.e. fluorescence) when the reaction occurs and is subsequently measured by a light sensor. We selected over a hundred natural and artificial proteins from the five lysozyme families for synthesis and evaluation. We use the activity assay to determine which proteins are properly functioning and to what level. Here’s what we discovered:
Artificial proteins work just as well as natural proteins
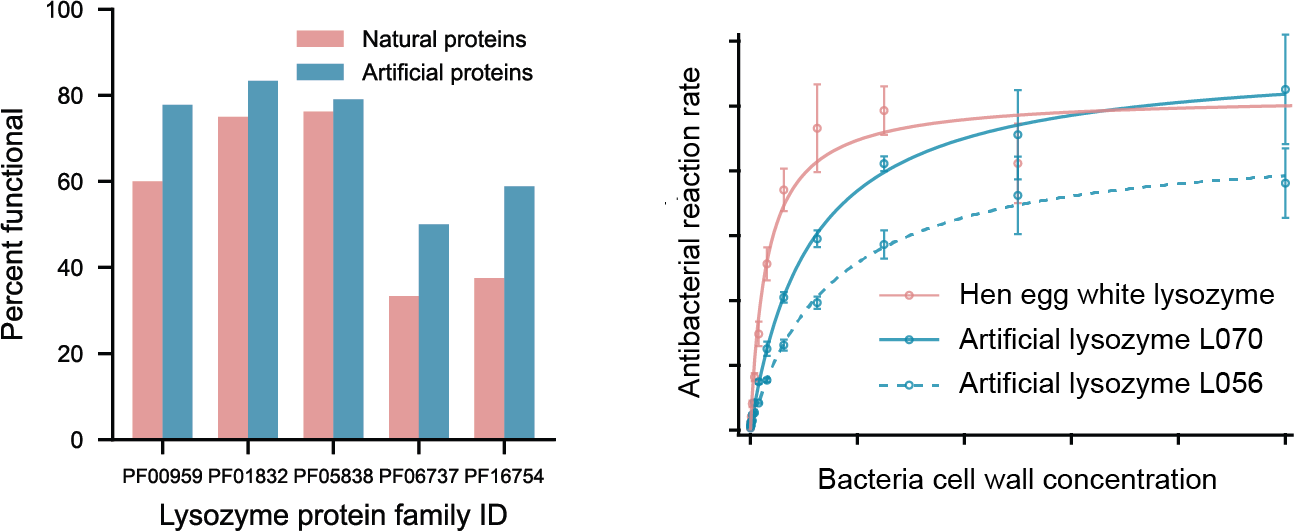
Among our artificial lysozymes, 73% were found to be functional antibacterial proteins, as compared to natural proteins which were 59% functional. Artificial proteins from all five evolutionary families of lysozyme showed activity.
To ensure the highest level of rigor, Professor Fraser’s lab conducted a gold-standard functional measurement (i.e. catalytic efficiency determination) of two of our artificially-designed lysozymes. The catalytic efficiencies for the two artificial lysozymes were comparable with hen egg white lysozyme, a highly functional antibacterial protein that has naturally evolved over many years.
Artificial proteins are highly diverse and unseen in nature
While one of our goals was to be able to generate functional artificial proteins, we also aimed to generate proteins that are very different from known natural proteins. Protein design algorithms typically take a known natural protein and change only a few amino acids to create an artificial protein. However, it is much more challenging to generate artificial proteins where a large fraction of acids are different from any known natural protein. We found that ProGen is able to do just that. For this analysis, we grouped our artificial proteins by how similar they are to the closest known natural protein in terms of amino acid occurrences and evaluated proteins that were 40-90% identical to natural proteins.
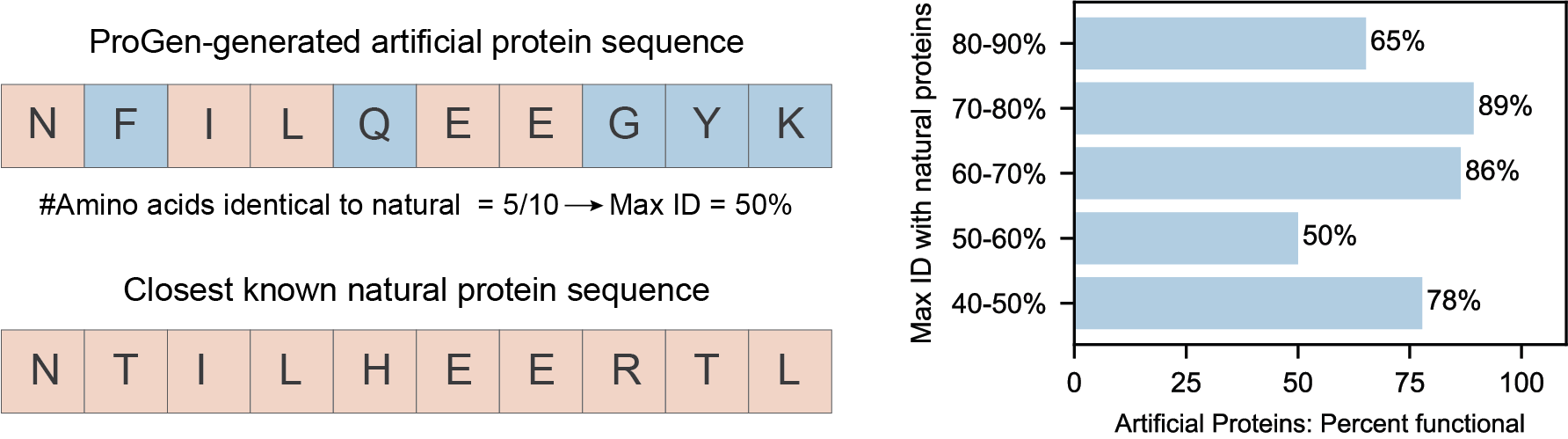
Artificial lysozymes in every similarity group between 40-90%-identity were functional, including lysozymes that were only 40-50% identical to any known protein in nature! In other words, ProGen was able to generate proteins that have up to 100 amino acids that are different from a 170-amino acid-long natural lysozyme sequence and can still retain antibacterial activity. This is akin to generating a paragraph of text that has more than half the words different than any human-written text, while still retaining the same, very precise, meaning.
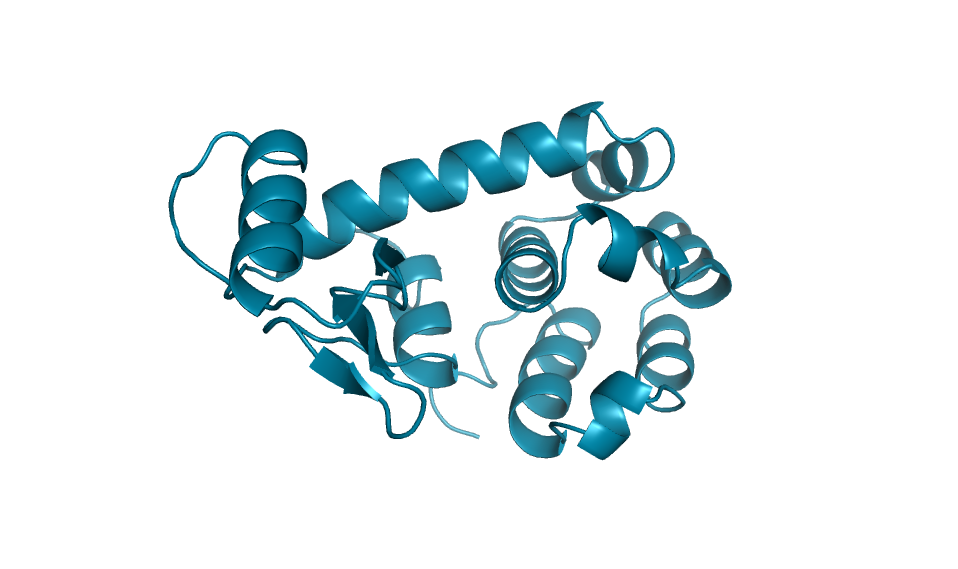
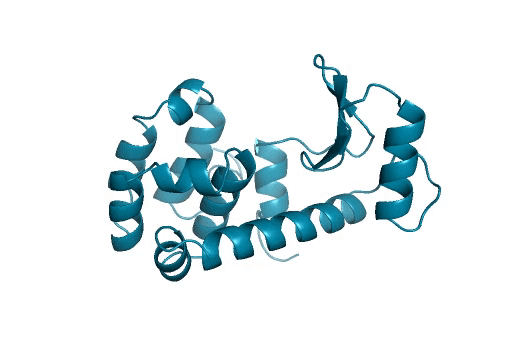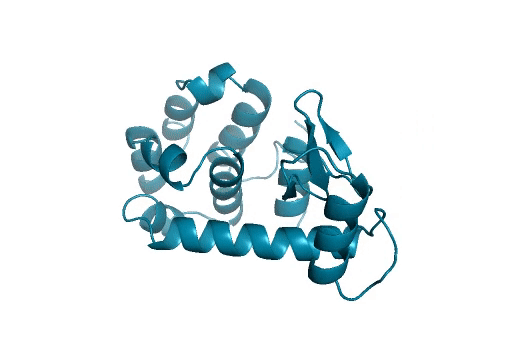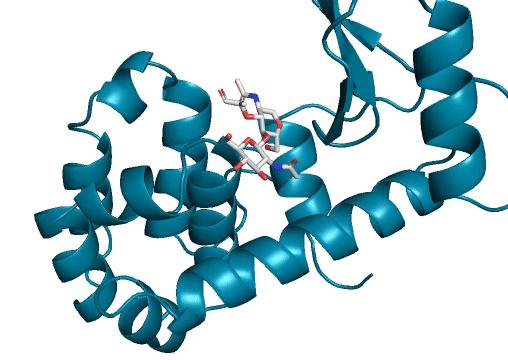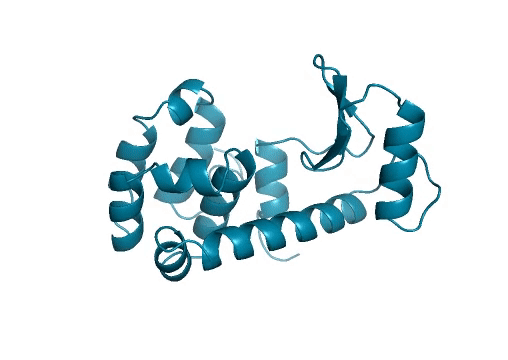
Marking history with a crystal structure
Scientists use techniques in X-ray crystallography to determine the structure of every atom in a protein. From this information, we can visualize the shape of a protein which is incredibly important for our understanding and advancement of protein science. Lysozymes were the first ever enzymes to have their structures determined by scientists. Likewise, we mark another first in our study. To the best of our knowledge, we have determined the first structure of a protein fully-designed by AI.
Towards A Universal Model for Artificial Protein Generation
With ProGen, we have developed a general-purpose AI model for generating novel proteins. In this work, we experimentally showed that ProGen-generated artificial antibacterial proteins are just as effective as natural proteins in killing bacteria while being unseen in nature. We also obtained the first known 3D structure of an AI-generated protein. Additional results covered in our paper show that ProGen can also accurately predict whether artificial proteins from two other very different protein families—chorismate mutase and malate dehydrogenase—are functional, based on experimental data from previous studies. Taken together, these experiments demonstrate that ProGen can generate functional artificial proteins from diverse protein families on demand.
In the near future, conditional generation of protein sequences could be used to design highly tailored proteins with desired properties, such as the ability to bind to another molecule or the ability to operate at high temperatures. Achieving these goals, with careful consideration of ethical implications, will allow us to quickly develop treatments for diseases or enzymes for industrial and environmental applications. More broadly, our work opens many new doors for utilizing state-of-the-art AI language modeling technology for accelerating protein engineering.
Authors
This study was an interdisciplinary effort led by Salesforce Research in collaboration with Tierra Biosciences and the Fraser Lab at UCSF. The study’s authors are Ali Madani, Ben Krause, Eric R. Greene, Subu Subramanian, Benjamin P. Mohr, James M. Holton, Jose Luis Olmos, Jr., Caiming Xiong, Zachary Z. Sun, Richard Socher, James S. Fraser, and Nikhil Naik. We thank many others within our respective institutions for their support and feedback. Please see our pre-print for a full list of acknowledgments. Feel free to contact the blog authors at {amadani,bkrause,nnaik}@salesforce.com.
Broader Impacts Statement
Finally, if ProGen or a future iteration thereof is adopted broadly, the use-cases for generated samples and their downstream effects should be carefully considered to ensure safe, non-nefarious, and ethical applications. For any technology that enables the discovery of new proteins, active oversight during project initiation, experimental optimization, and deployment phases should be put in place to ensure safe usage and limitation of unintended harmful effects.
