Learning to Extrapolate with Generative AI Models
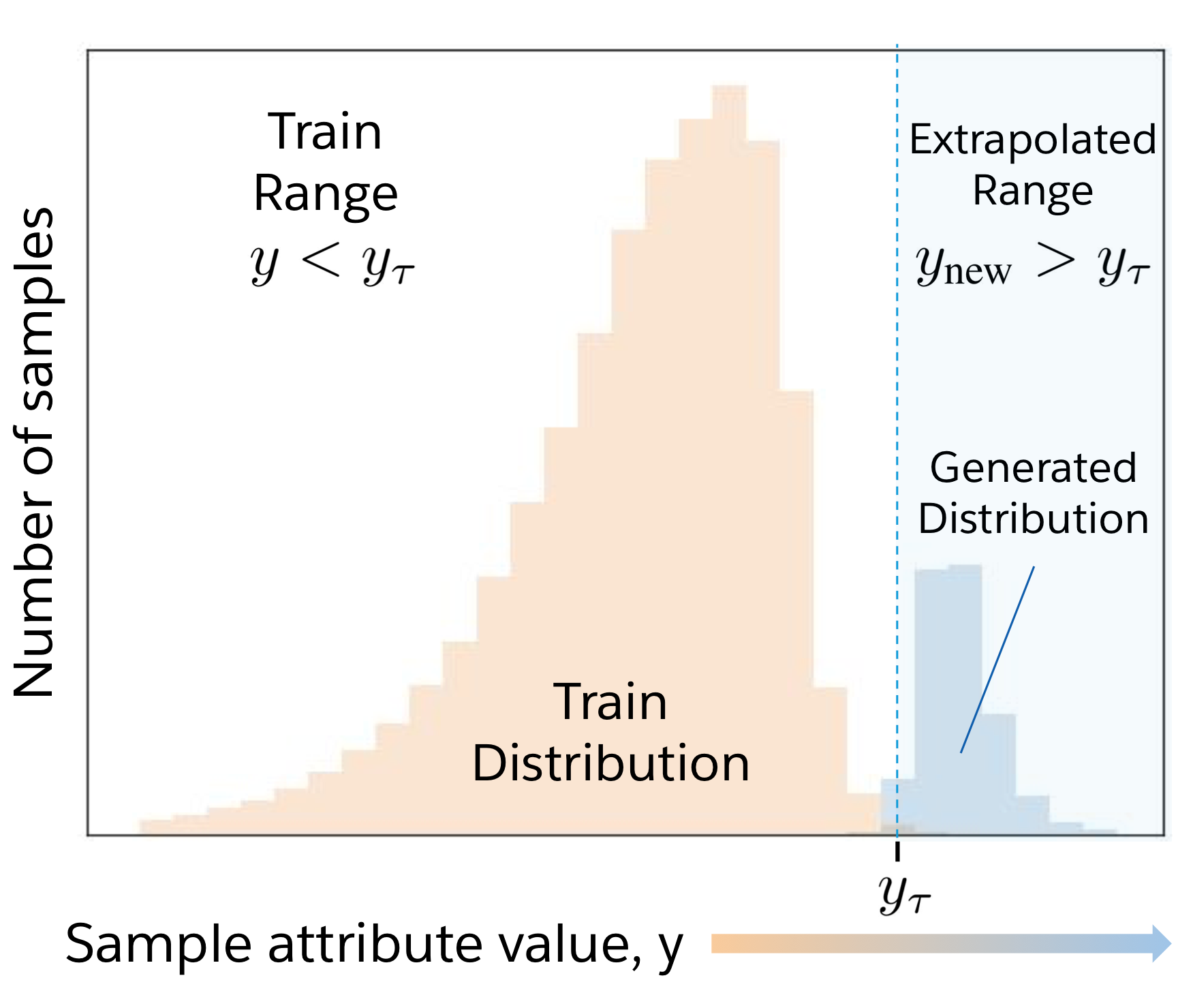
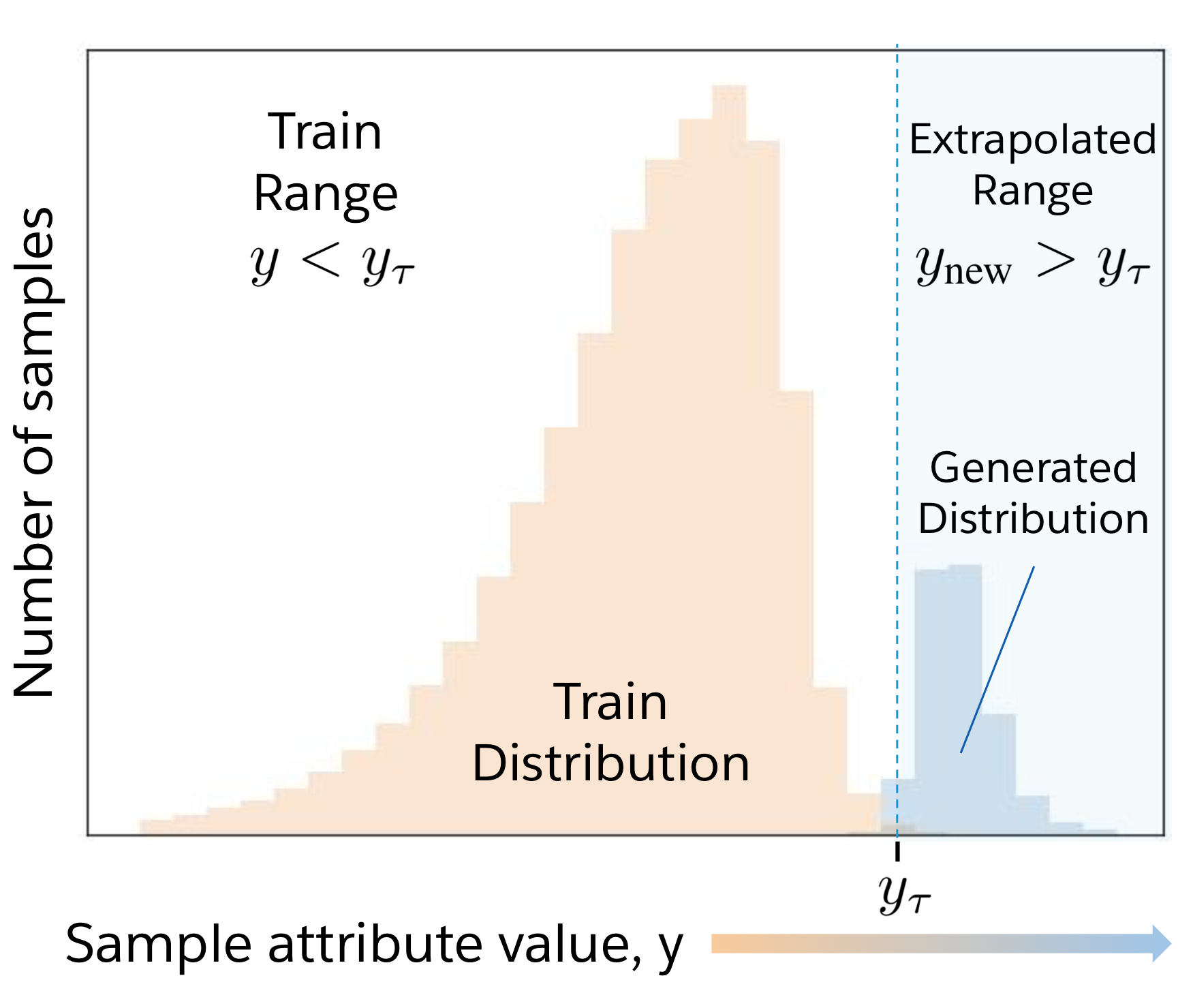
Deep generative models can generate impressively realistic text and images. In the physical sciences, synthetic design of proteins and molecules would greatly benefit from the ability to generate samples that improve upon a property of interest, which we term "attribute-enhanced generation". Deep generative models may provide a solution but struggle when operating beyond their training distribution. In this work, we develop GENhance, an encoder-decoder model with a controllable latent space, that can extrapolate to generate strongly-positive movie reviews and highly-stable protein sequences without ever being exposed to similar data in training. Extrapolation with generative AI models could have far-reaching implications for biology, chemistry, and natural language processing. Please refer to our paper for details.
Summary: An AI model can generate sequences across natural language and proteins with attributes that go beyond the training distribution.
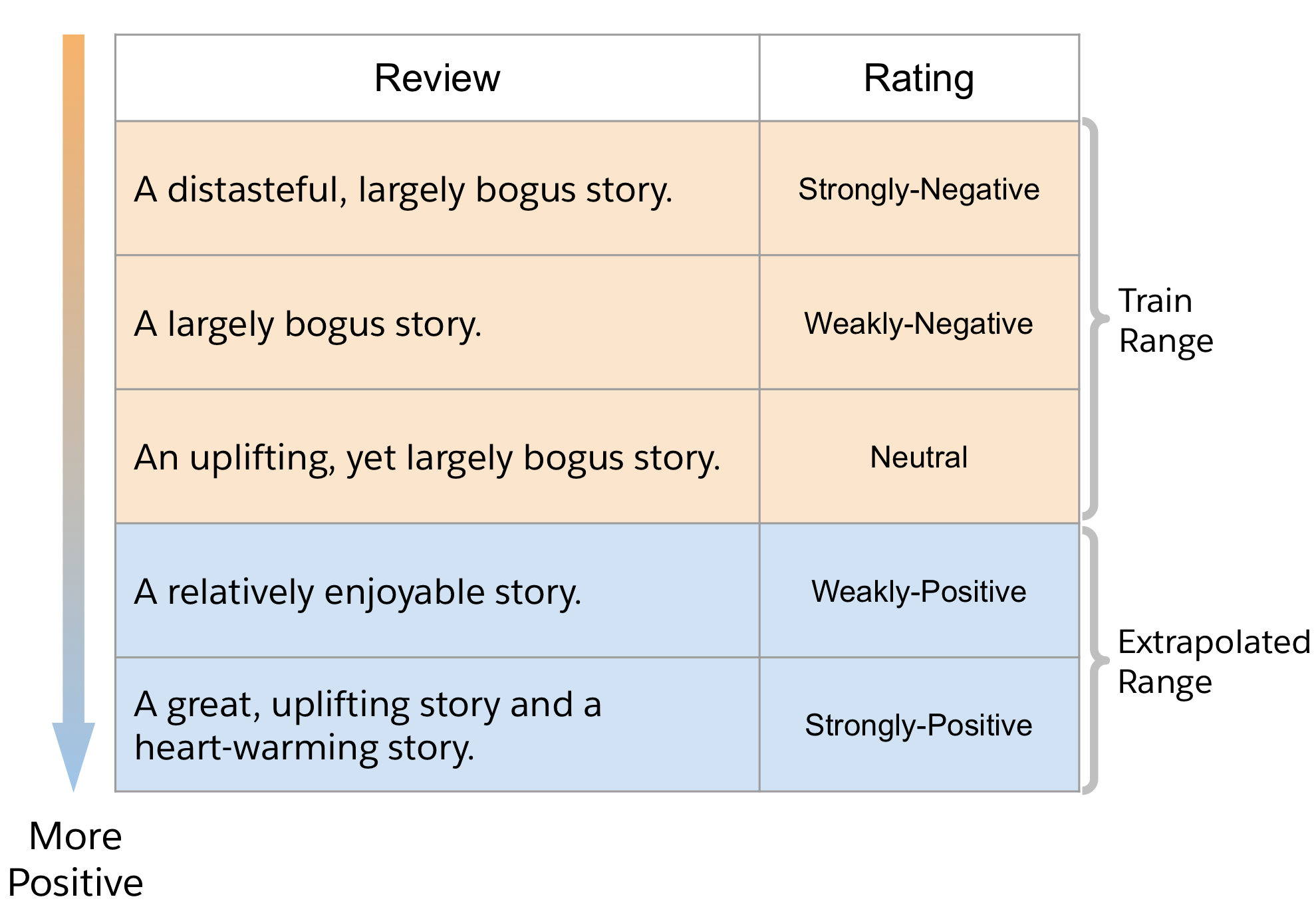
A Natural Language Example: Movie Reviews Sentiment
Let’s look at a movie review dataset, SST-5 [1]. It contains movie reviews each with an associated label for sentiment ranging from 1 to 5: strongly-negative, weakly-negative, neutral, weakly-positive, and strongly-positive. Imagine you were *only* exposed to negative and neutral movie reviews in supervised training, how well would you expect to generate a movie review with a positive sentiment?
Extrapolation with GENhance
To generate movie reviews with positive sentiment, we develop a framework that consists of a generator and a discriminator. The generator would synthesize reviews with enhanced positivity while the discriminator would subsequently rank the reviews by sentiment. A key feature of the generator is a mechanism for sentiment control for synthesized reviews. This is performed by manipulating a review’s latent vector, which is formed by passing the review text into GENhance’s encoder module in a compression process. This vector is a compact representation of the text that may encode language attributes such as content, syntax, tone and sentiment. We can alter the attributes of the original review with specified perturbations to the latent vector. By editing the portion of the vector that encodes for the sentiment of the text, we can increase the positivity. Passing the edited vector through GENhance’s decoder would decompress it to generate a new movie review with enhanced sentiment.
We use a combination of training objectives critical to the success of GENhance. For generated reviews to be as fluent as the training samples, we train the model with a reconstruction objective to recreate an original input review from its latent vector. To encode sentiment in the compressed vector form, we train the model to correctly rank the positivity of two review texts that are randomly sampled from different classes through their latent vectors. With an additional objective that smoothens the space of encoded vectors, GENhance’s generated reviews can still look realistic even when their vectors are altered for increased positiveness.
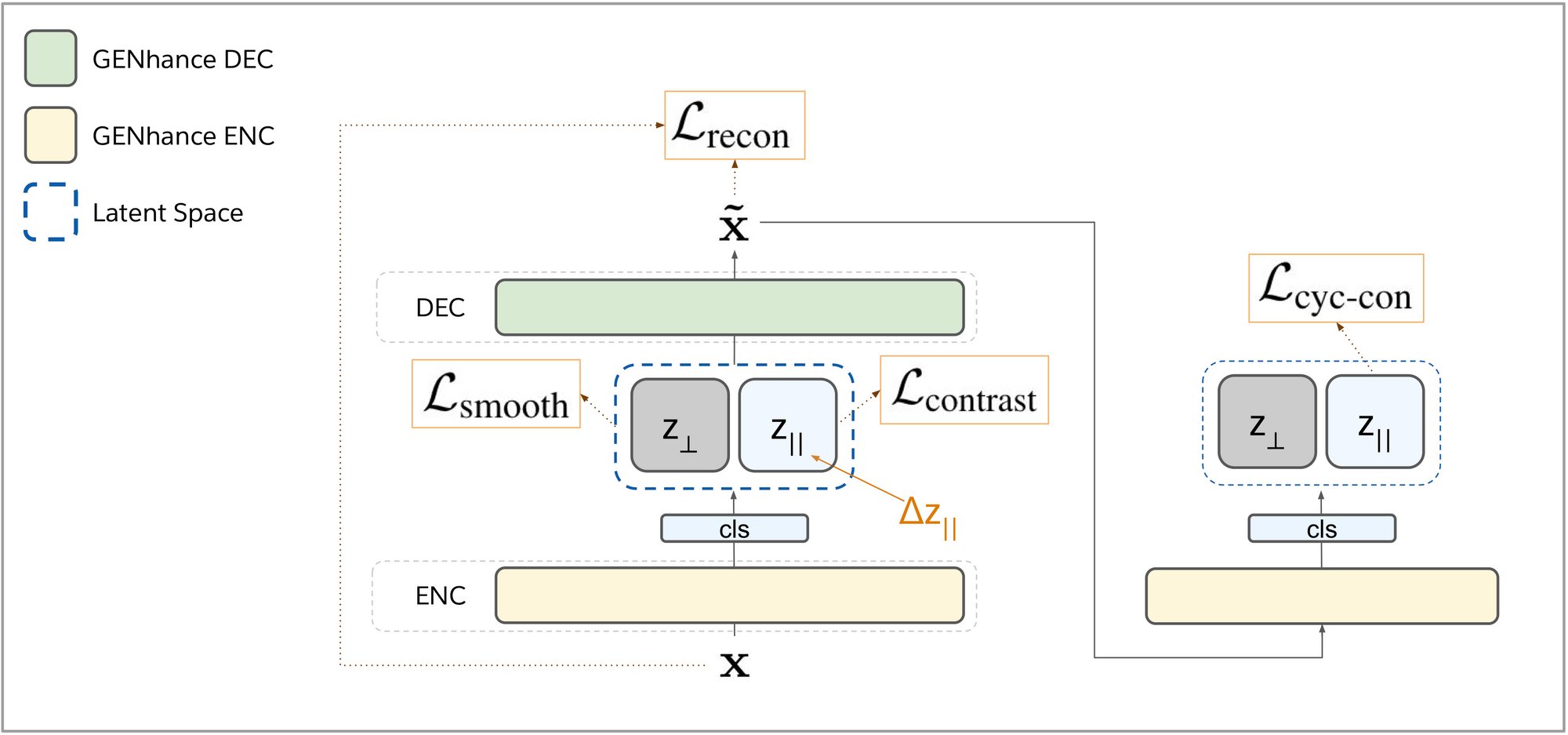
Remember that we would still need a discriminator model to help us rank generated reviews so that we can find the ones with the highest positive sentiment? Since the encoder module was already trained to correctly rank sequences based on their attribute of interest (e.g. positivity), we can also use it as our discriminator. As the encoder was only trained on human written review text, using it directly to rank synthetic reviews might have poor accuracy due to an out-of-distribution effect. To familiarize the encoder with synthetic text, we design a cycle-consistency objective which trains it to correctly rank reconstructed review texts. Relying on the success of the pretraining paradigm, we initialized the weights of GENhance with a pretrained T5 language model [2].
Extrapolating to Positive Movie Reviews
To evaluate GENhance’s performance in enhancing positive sentiment, we use neutral-labeled reviews as inputs and alter their vectors in the “positive direction” before generating new reviews with the decoder. Some examples are shown below:
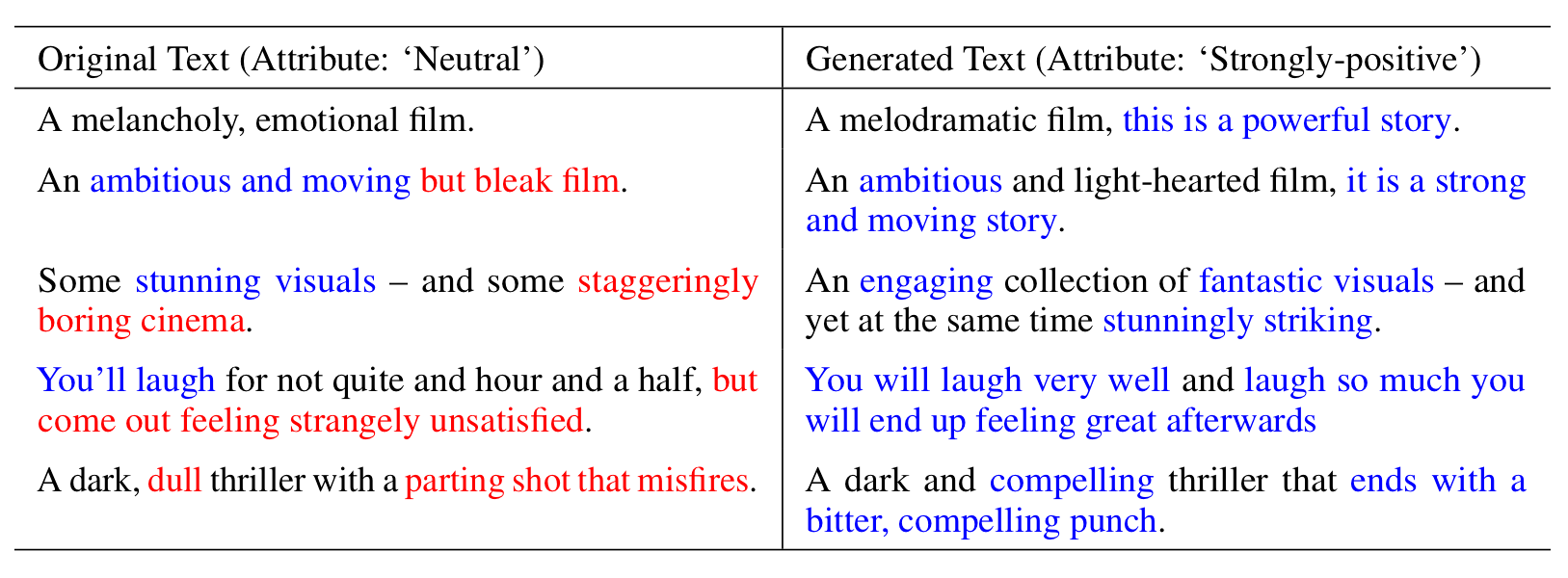
We also compared GENhance with two types of baselines. The first (Baseline Gen-Disc) uses similar generator and discriminator modules as GENhance but without a latent space inside the generator or the ability to condition original text. The second baseline (MCMC-T5) iteratively replaces text spans in reviews guided by T5, a pretrained language model. GENhance improves the positiveness of the review text with a larger fraction of positive review text than these baselines. Our experiments also show that the smoothing and cycle-consistency training objectives are important for GENhance performance.

Engineering Stable ACE2 Subdomains
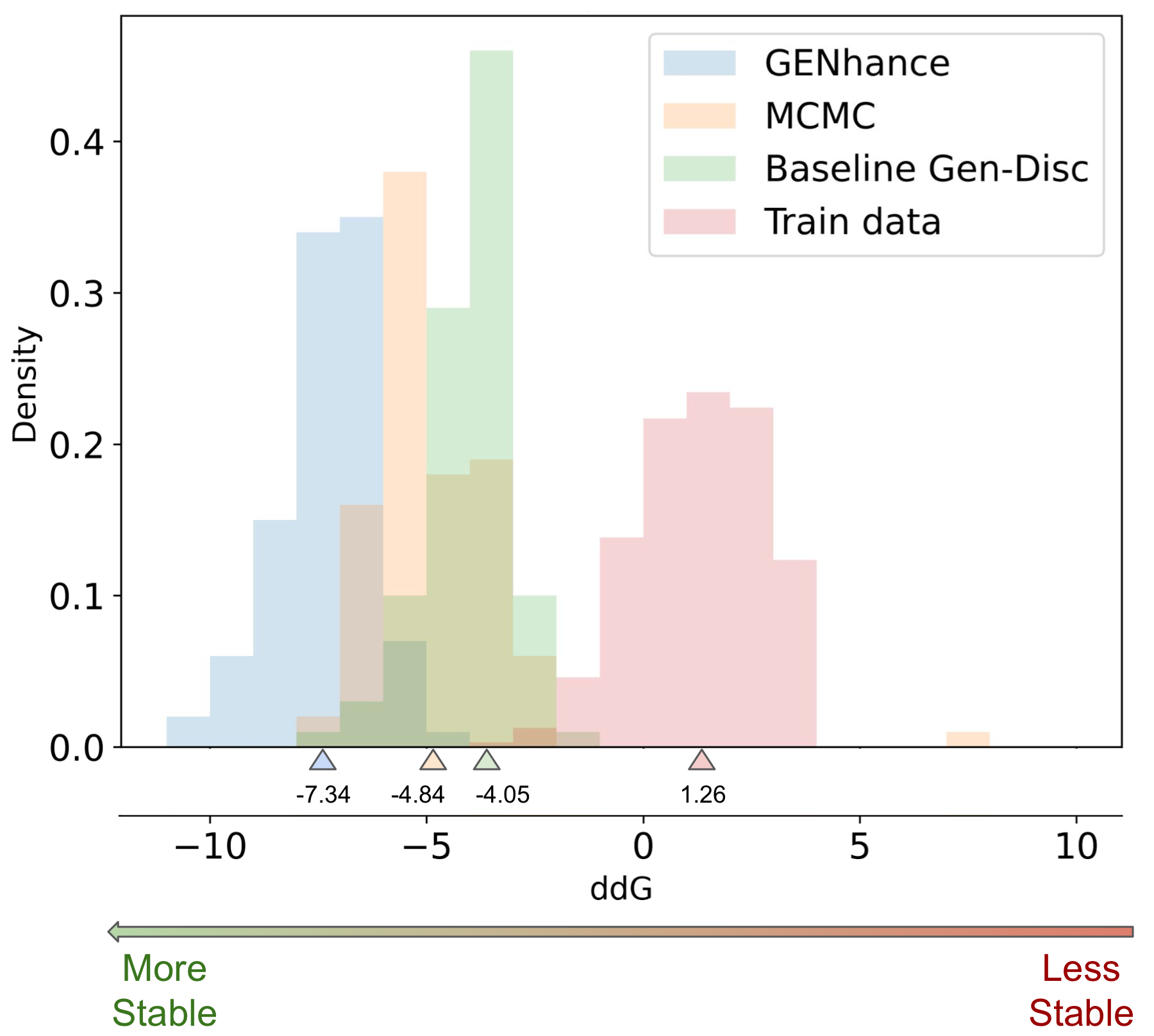
Designing a protein with an optimized property (e.g. stability) is of immense interest in the fields of synthetic biology and drug discovery. Since the SARS-CoV-2 virus binds to ACE2 to gain entry into human organs, ACE2 has emerged as a promising target for COVID-19 therapeutic protein design [3]. We used GENhance to train on a dataset of ACE2 mutants and their stability labels, computed as change in free energy, ddG, between the folded and unfolded state of a protein sequence with the known ACE2 structure. The aim is to generate ACE2-like protein sequences that are more stable, i.e. with a lower ddG value, than anything seen before.
Our extrapolation results look as impressive in the protein domain as in natural language. The distribution of ACE2 variants generated from GENhance have lower/improved mean and max stability (ddG) as compared to the training distribution and baseline methods.
Future Implications
Many of the transformative solutions in biology and chemistry will come from discovering proteins, materials, or molecules with optimized properties-- beyond what currently exists in nature. With GENhance [5] we take the first step towards this goal with deep learning by tackling extrapolation in sequence generation. In addition, GENhance may also be applied to language modeling for reducing toxicity or improving desirable attributes in AI-generated natural language. We release our code along with all models and data in hopes of further investigation of generative modeling extrapolation and data-driven design.
Authors
The first authors of the work are Alvin Chan, who was an intern as Salesforce Research, and Ali Madani. Other authors include Ben Krause and Nikhil Naik.
Many thanks to Nitish Shirish Keskar and other members of the Salesforce Research team for feedback and comments.
References
[1] Richard Socher, Alex Perelygin, Jean Wu, Jason Chuang, Christopher D Manning, Andrew Y Ng, and Christopher Potts. Recursive deep models for semantic compositionality over a sentiment treebank. In Proceedings of the 2013 conference on empirical methods in natural language processing, pp. 1631–1642, 2013.
[2] Colin Raffel, Noam Shazeer, Adam Roberts, Katherine Lee, Sharan Narang, Michael Matena, Yanqi Zhou, Wei Li, and Peter J Liu. Exploring the limits of transfer learning with a unified text-to-text transformer. arXiv preprint arXiv:1910.10683, 2019.
[3] Kui K Chan, Danielle Dorosky, Preeti Sharma, Shawn A Abbasi, John M Dye, David M Kranz, Andrew S Herbert, and Erik Procko. Engineering human ace2 to optimize binding to the spike protein of sars coronavirus 2. Science, 369(6508):1261–1265, 2020.
[4] Schymkowitz, J., Borg, J., Stricher, F., Nys, R., Rousseau, F., & Serrano, L. (2005). The FoldX web server: an online force field. Nucleic acids research, 33(suppl_2), W382-W388.
[5] Alvin Chan, Ali Madani, Ben Krause, Nikhil Naik. Deep Extrapolation for Attribute-Enhanced Generation. arXiv preprint arXiv:2107.02968, 2021.
